publications
* denotes equal contribution
An up-to-date list is available on Google Scholar.
2022
-
 ×Evaluation of thermodynamic consistency of kinetic parameters in cyclic enzyme-catalyzed reaction networksSarang S. NathChemical Physics Letters, 2022
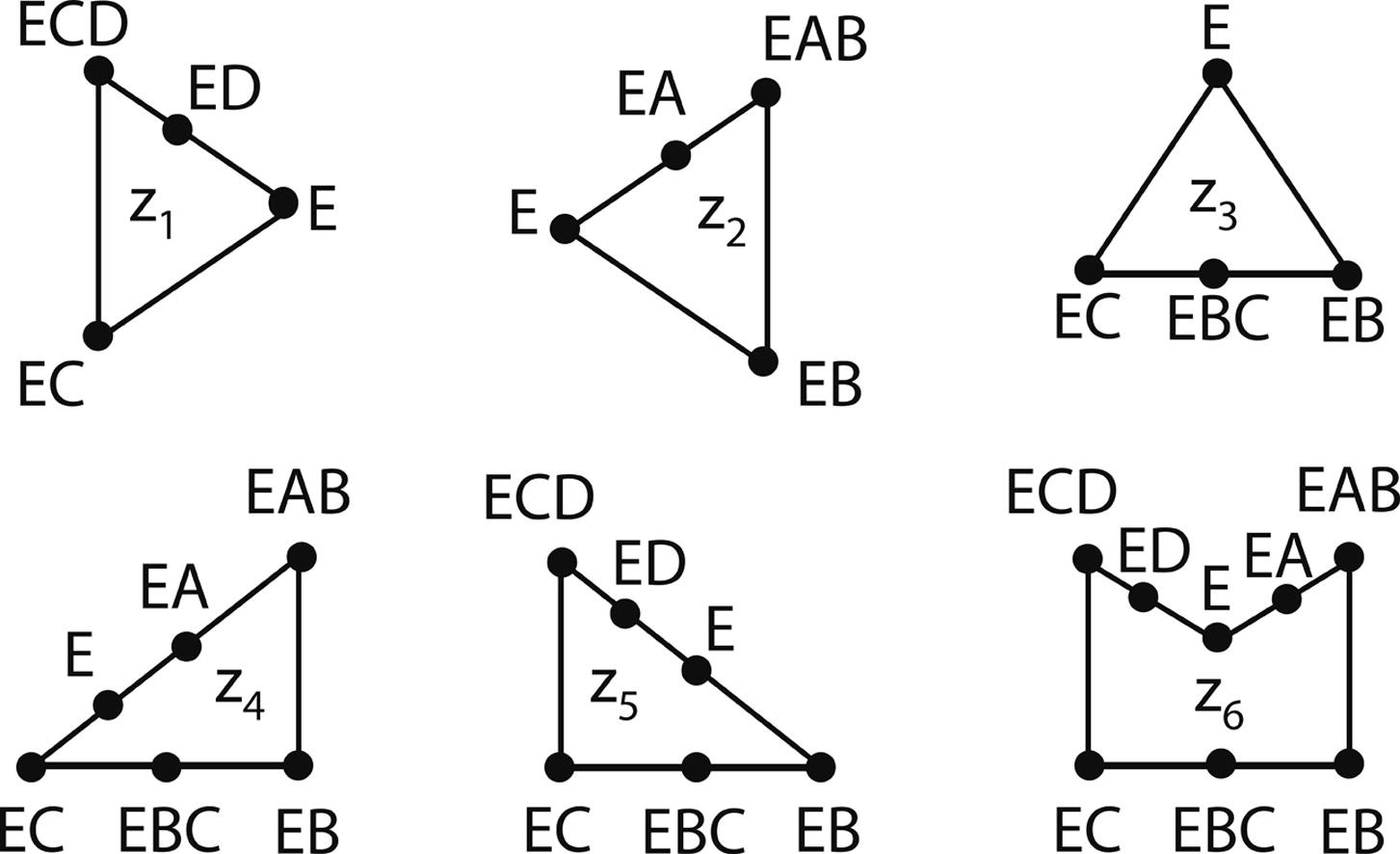
×Evaluation of thermodynamic consistency of kinetic parameters in cyclic enzyme-catalyzed reaction networksSarang S. NathChemical Physics Letters, 2022It is shown by analysis of a cyclic enzymatic network that the presence of zero net flux/current cycles impose strong thermodynamic constraints on the values of the kinetic parameters. Thermodynamic analysis is carried out for the four-node pyramidal reaction network involving the conversion of 7,8-dihydrofolate (H2F) and NADPH in the presence of H+ to 5,6,7,8-tetrahydrofolate (H4F) and NADP+ catalyzed by the universal enzyme dihydrofolate reductase (DHFR). The complete set of empty reaction routes in the DHFR pathway is identified, equations for the thermodynamic constraints imposed on the rate constants of the various elementary steps are written down, and the thermodynamic consistency of the experimentally determined rate constants is evaluated. The visual approach based on the theory of graphs can be employed to test the thermodynamic consistency of kinetic parameters of general cyclic enzyme-catalyzed reaction networks.
-
 ×Elucidating dynamics and mechanism of cyclic bioreaction networks using topologically-equivalent electrical circuitsSarang S. Nath, Lars K. Nielsen, and John VilladsenChemical Engineering Science, 2022
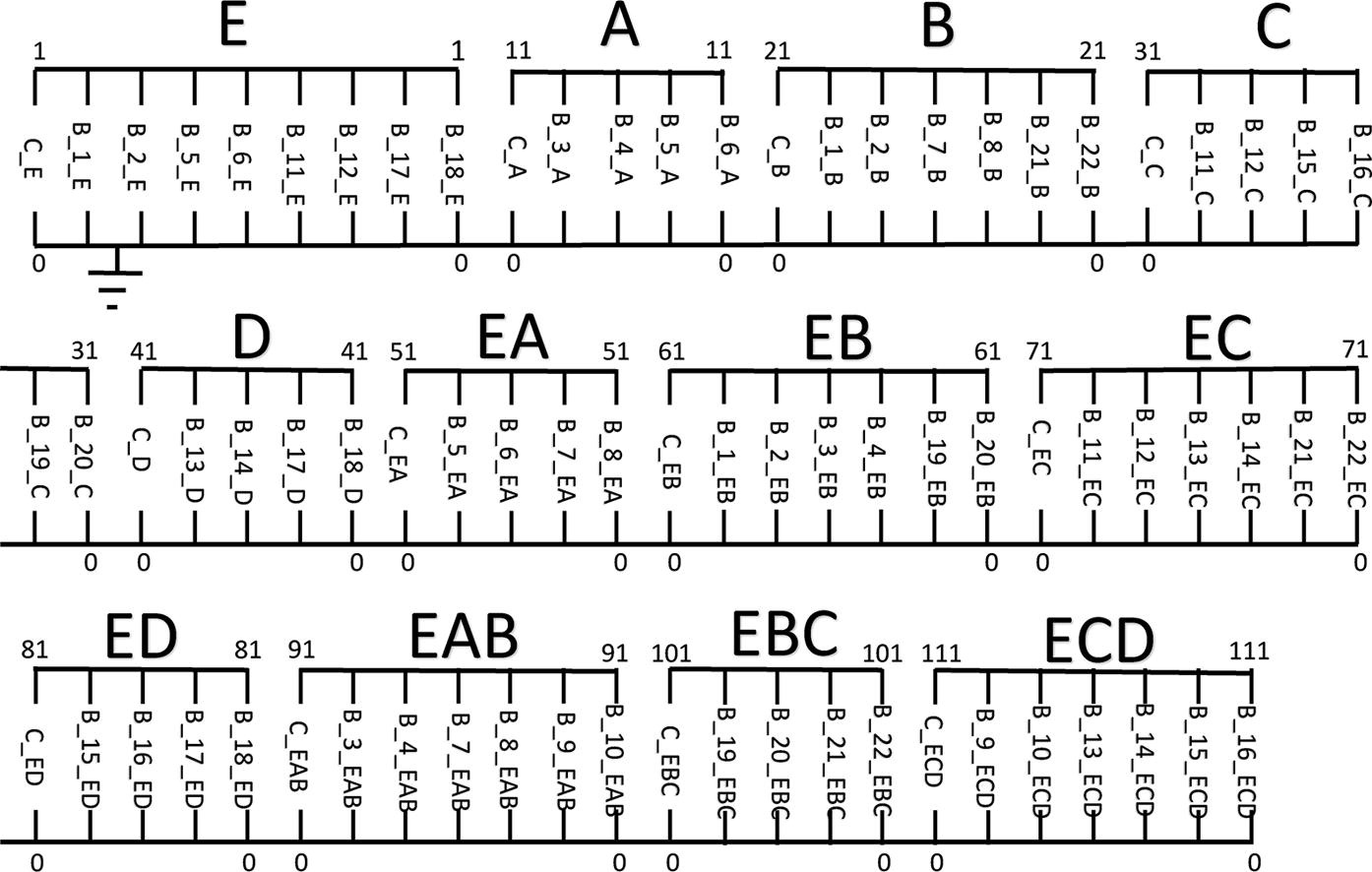
×Elucidating dynamics and mechanism of cyclic bioreaction networks using topologically-equivalent electrical circuitsSarang S. Nath, Lars K. Nielsen, and John VilladsenChemical Engineering Science, 2022A novel methodology is established to convert a set of elementary chemical reactions into topologically-equivalent electrical circuits. The circuit analog is then simulated to elucidate the dynamics of complex cyclic bionetworks. The pyramidal reaction network catalyzed by the universal enzyme dihydrofolate reductase – which has important pharmacological and medical relevance in connection with cancer therapy – is used to illustrate the application of our model. The finite number of mechanisms, sub-cycles, or graphs constituting the cyclic network are analyzed, their contribution to the overall steady-state reaction rate determined, and the most probable mechanism is identified. The developed methodology is elegant, modular, and permits representation and visualization of network structure and topology at a glance. It can handle linear and nonlinear kinetics/rate laws, does not require simplifying assumptions such as use of the quasi-steady-state/Bodenstein approximation or the absence of nonlinear kinetic steps in the intermediates, and is suitable for coupling and integration with other flux-based reaction models.
-
 ×Modeling dynamics of chemical reaction networks using electrical analogs: Application to autocatalytic reactionsSarang S. Nath, and John VilladsenChemical Engineering Journal Advances, 2022
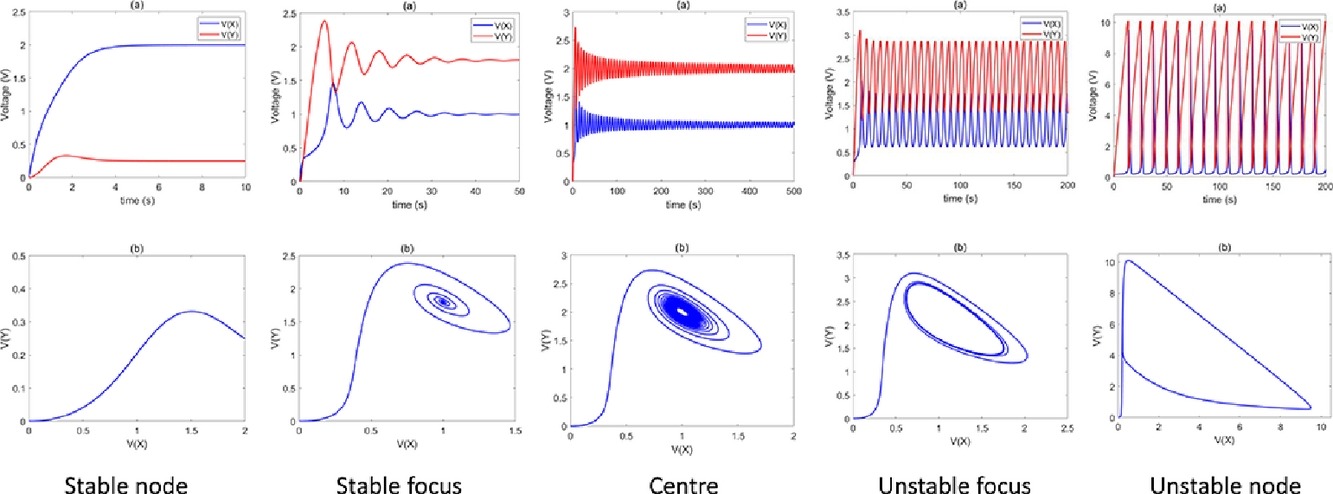
×Modeling dynamics of chemical reaction networks using electrical analogs: Application to autocatalytic reactionsSarang S. Nath, and John VilladsenChemical Engineering Journal Advances, 2022Modeling complex chemical reaction networks has inspired a considerable body of research and a variety of approaches to modeling nonlinear pathways are being developed. Here, a general methodology is formulated to convert an arbitrary reaction network into its equivalent electrical analog. The topological equivalence of the electrical analog is mathematically established for unimolecular reactions using Kirchhoff’s laws. The modular approach is generalized to bimolecular and nonlinear autocatalytic reactions. It is then applied to simulate the dynamics of nonlinear autocatalytic networks without making simplifying assumptions, such as use of the quasi-steady state/Bodenstein approximation or the absence of nonlinear steps in the intermediates. This is among the few papers that quantify the dynamics of a nonlinear chemical reaction network by generating and simulating an electrical network analog. As a realistic biological application, the early phase of the spread of COVID-19 is modeled as an autocatalytic process and the predicted dynamics are in good agreement with experimental data. The rate-limiting step of viral transmission is identified, leading to novel mechanistic insights.
-
 ×Minimal model for oscillatory dynamics of a nonlinear chemical reaction networkSarang S. NathPhysica D: Nonlinear Phenomena, 2022
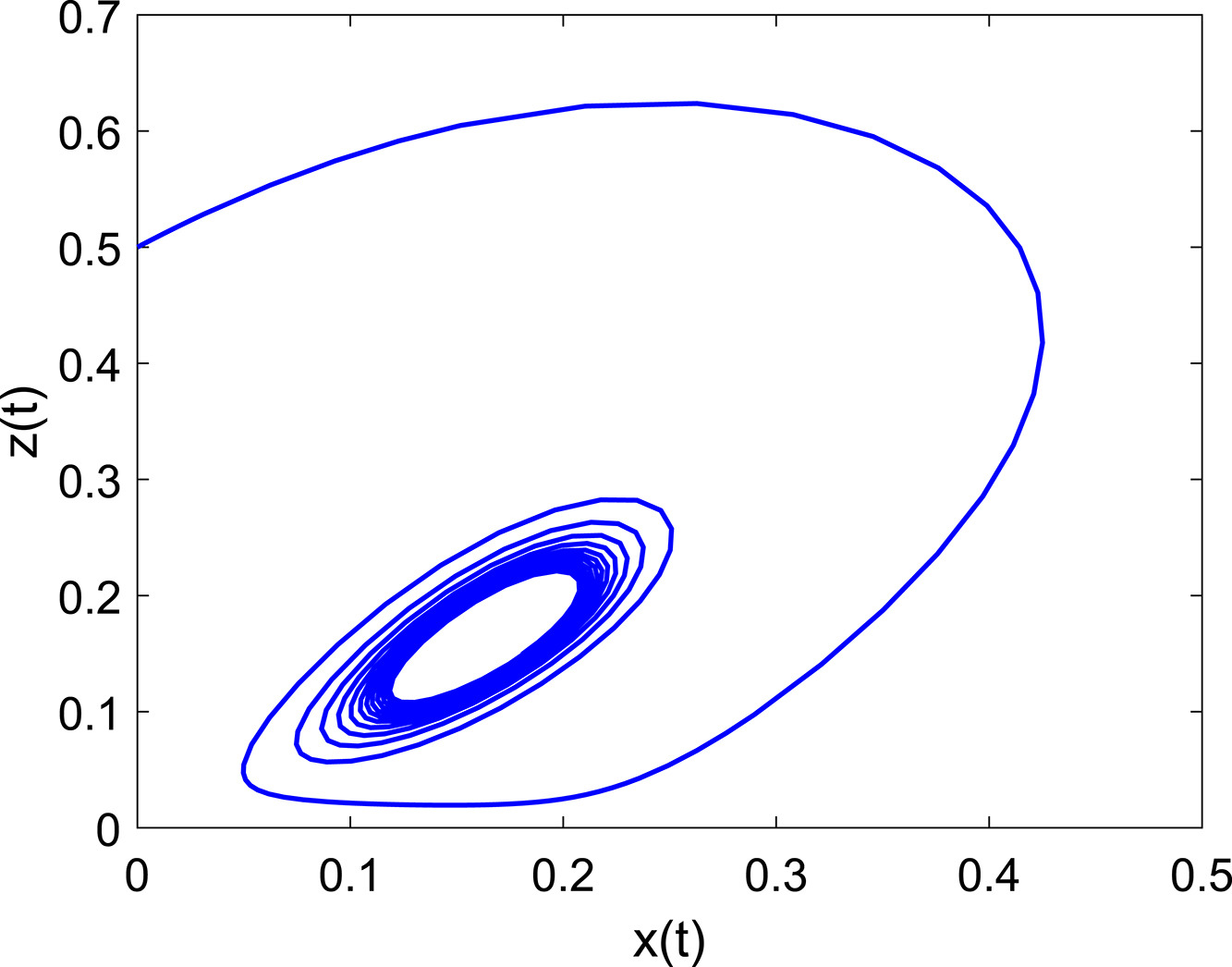
×Minimal model for oscillatory dynamics of a nonlinear chemical reaction networkSarang S. NathPhysica D: Nonlinear Phenomena, 2022Since Bray’s pioneering discovery exactly 100 years ago of the first oscillating reaction, more than 25 families of nonlinear oscillating reaction networks have been identified. The B–Z reaction network has been expanded, and complex models considering a score of chemical reactions and species have been developed. Here we look at the inverse problem and ask how the complexity of the models can be reduced. We quantify the dynamics of a minimal model by linear stability techniques and direct integration of the set of ODEs, and show that essential oscillatory and excitability features are satisfactorily captured. Reasons for the success of the minimal model are explained. Extensions of the model to fundamental chemical and biological processes are discussed, and application of the chemical reaction network to mathematically model nerve conduction is illustrated.
-
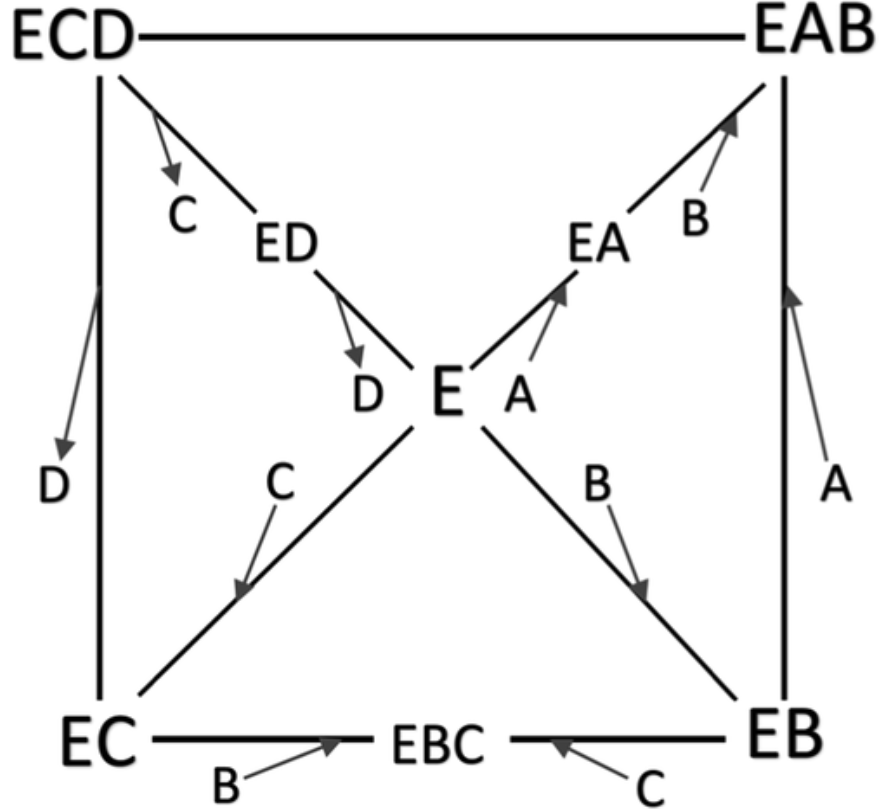 ×Thermodynamic Consistency and Kinetic Mechanism of a Complex Reaction Pathway by Analysis of Its Constituent Cyclic NetworksSarang S. NathIndustrial & Engineering Chemistry Research, 2022
×Thermodynamic Consistency and Kinetic Mechanism of a Complex Reaction Pathway by Analysis of Its Constituent Cyclic NetworksSarang S. NathIndustrial & Engineering Chemistry Research, 2022The complete set of thermodynamically consistent sub-cyclic routes that produce a net positive reaction flux in the dihydrofolate reductase (DHFR) network is identified by a novel approach based on the theory of graphs. The net positive flux cycles that yield the minimal set of linearly independent rate equations (“mechanisms”) are then selected. The steady-state flux (reaction rate) through these constituent sub-cycles is quantified, and the sub-cycle that generates the maximum flux is identified as the most probable mechanism of the pathway. This analysis is performed for both wild-type and mutant Escherichia coli DHFR systems. The graph-theoretical method of visualization of networks developed here should prove to be a useful tool for elucidating the kinetic mechanism complex cyclic reaction networks.
2013
-
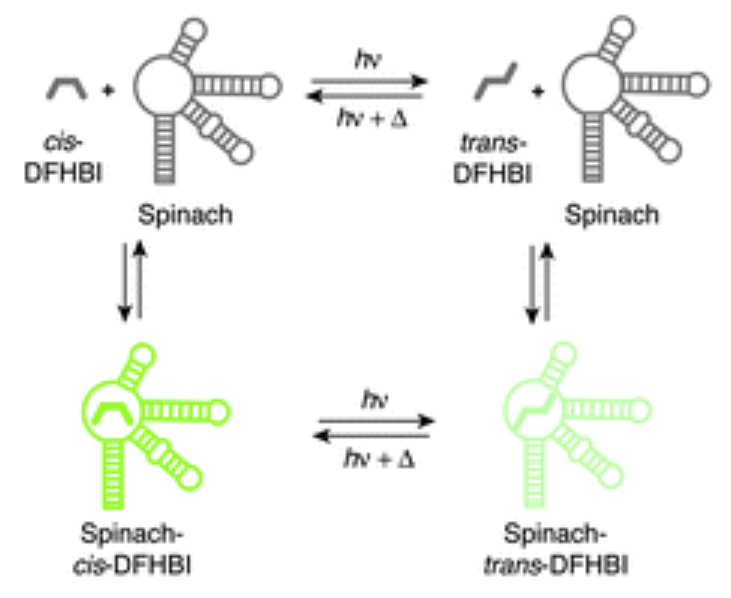 ×Photochemical properties of Spinach and its use in selective imagingPengcheng Wang, Jérôme Querard, Sylvie Maurin, Sarang S. Nath, Thomas Le Saux, Arnaud Gautier, and Ludovic JullienChem. Sci., 2013
×Photochemical properties of Spinach and its use in selective imagingPengcheng Wang, Jérôme Querard, Sylvie Maurin, Sarang S. Nath, Thomas Le Saux, Arnaud Gautier, and Ludovic JullienChem. Sci., 2013The progress in imaging instrumentation and probes has revolutionized the way biologists look at living systems. Current tools enable both observation and quantification of biomolecules, allowing the measurement of their complex spatial organization and the dynamic processes in which they are involved. Here, we report reversible photoconversion in the Spinach system, a recently described fluorescent probe for RNA imaging. Upon irradiation with blue light, the Spinach system undergoes photoconversion to a less fluorescent state and fully recovers its signal in the dark. Through thermodynamic titration, stopped-flow, and light-jump experiments, we propose a dynamic model that accounts for the photochemical behavior of the Spinach system. We exploit the dynamic fluorogen exchange and the unprecedented photoconversion properties in a non-covalent fluorescence turn-on system to significantly improve signal-to-background ratio during long-term microscopy imaging.
2009
-
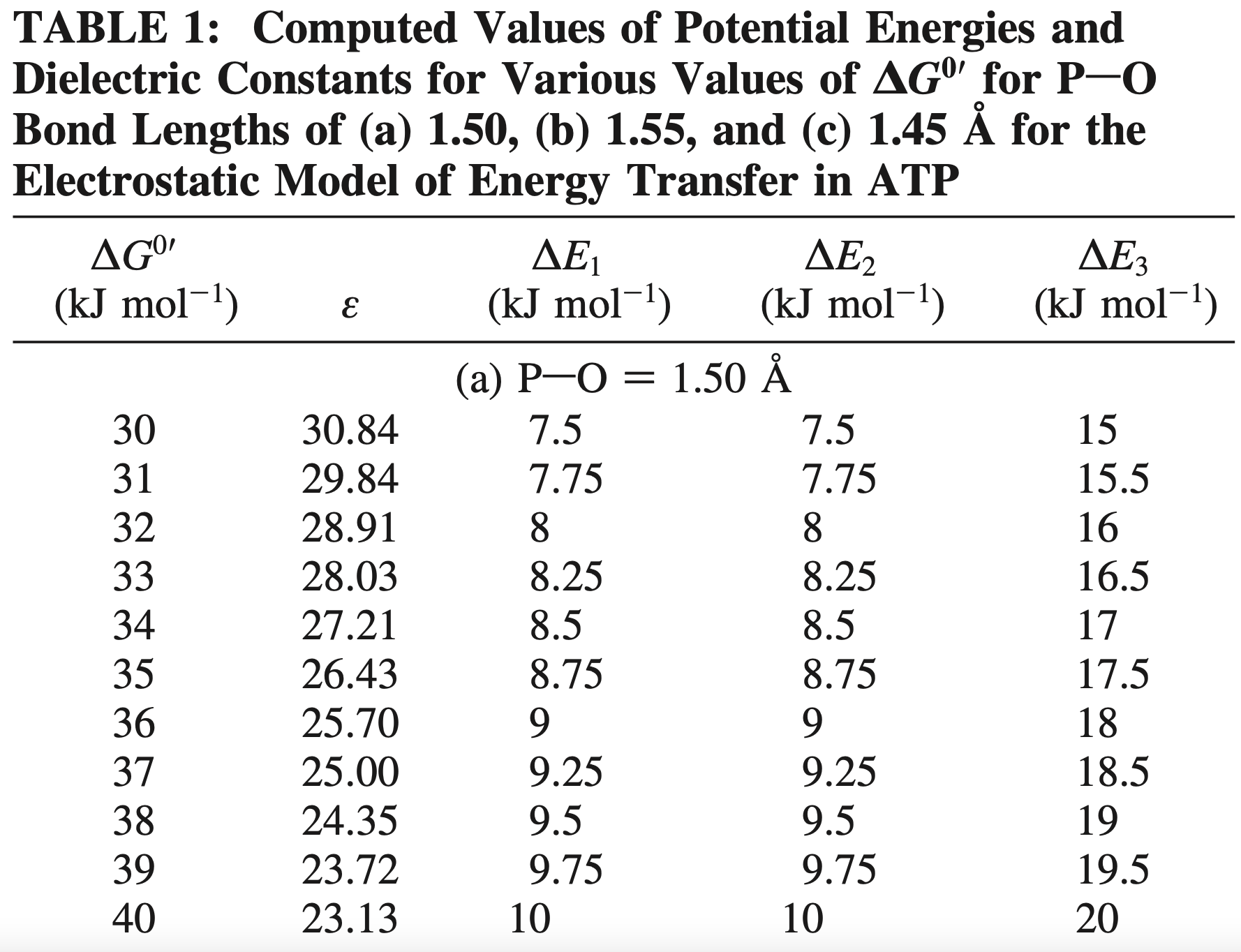 ×Energy Transfer from Adenosine Triphosphate: Quantitative Analysis and Mechanistic InsightsSarang S. Nath, and Sunil NathThe Journal of Physical Chemistry B, 2009
×Energy Transfer from Adenosine Triphosphate: Quantitative Analysis and Mechanistic InsightsSarang S. Nath, and Sunil NathThe Journal of Physical Chemistry B, 2009The ATP−ADP thermodynamic cycle is the fundamental mode of energy exchange in oxidative phosphorylation, photophosphorylation, muscle contraction, and intracellular transport by various molecular motors and is therefore of vital importance in biological energy transduction and storage. Following a recent suggestion in the pages of this journal (Ross, J. J. Phys. Chem. B 2006, 110, 6987−6990), we have carried out a simple quantitative analysis of a direct molecular mechanism of energy transfer from adenosine triphosphate (ATP). The simulation provides new insights into the mechanistic events following terminal phosphorus−oxygen bond cleavage during ATP hydrolysis. This approach also allows for the division of the energy-transfer process into elementary steps and for the prediction of the distribution of the standard-state Gibbs free energy of the overall ATP hydrolysis process among the various steps of substrate binding, bond cleavage, and product release in the enzymatic cycle, which had proved very difficult to specify previously. These predictions are consistent with available experimental data on ATP hydrolysis by protein biomolecular machines. The fundamental biological implications arising from our results are also discussed in detail. The aspects considered in this work enable us to look at the entire process of ATP synthesis/hydrolysis and energy transduction and storage in various biological molecular machines in a logically consistent and unified way.